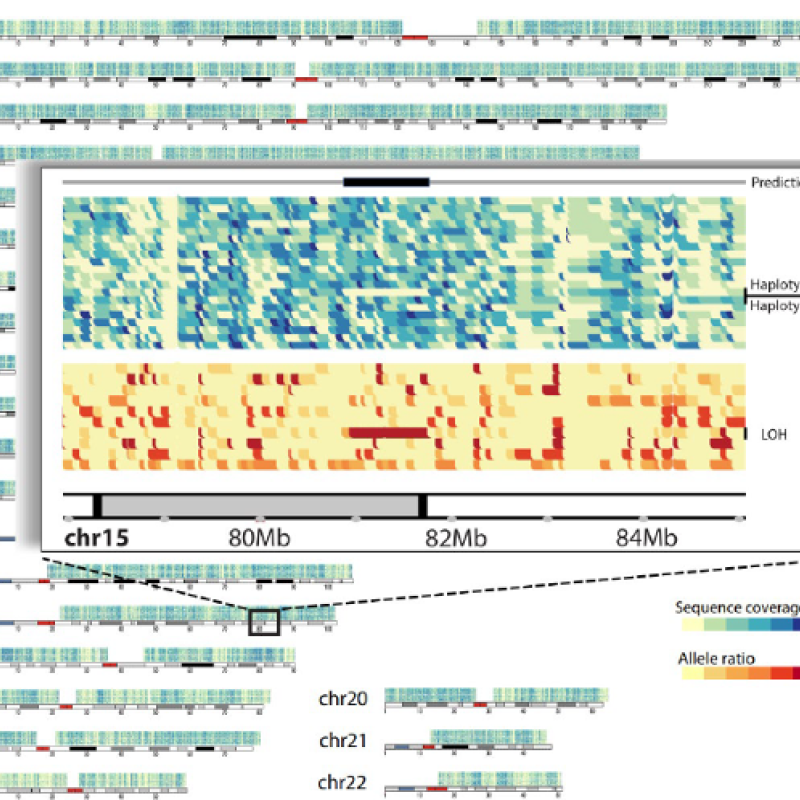
Meet Kelly Sovacool, a Ph.D. candidate in DCMB and in Pat Schloss’s lab (Microbiology and Immunology)
Kelly Sovacool will defend her Ph.D. in Bioinformatics, June 22nd, 2023. The title of her dissertation is “Improving machine learning models for microbiome analysis and democratizing data science along the way.”